Translate this page into:
Why we should rely only on molecular tests based on Indian data

*Corresponding author: Purvish M. Parikh, Group Oncology Director Academic, Shalby Cancer and Research Institute, Mumbai, Maharashtra, India. purvish1@gmail.com
-
Received: ,
Accepted: ,
How to cite this article: Parikh PM, Singh R. Why we should rely only on molecular tests based on Indian data. Int J Mol Immuno Oncol 2020;5(2):44-6.
Over the past 15 years, lung cancer has become the poster boy of personalized medicine, targeted therapy, and immuno-oncology.[1] Gefitinib initiated the transformation of how we treat lung cancer today, a great example of a drug that would have been lost in oblivion but for the effort to find out ethnic differences in molecular mutations.[2,3] Not only are there genetic differences between races (Caucasians vis-a-vis Asians), there is also heterogeneity within a single country like India.[4] Moreover, such variations have clinical implications in personalized treatment choices for optimizing patient management.[5] Figure 1 shows how such a systematic approach in sequencing therapy can lead to significant improvement in the survival of metastatic adenocarcinoma of the lung.
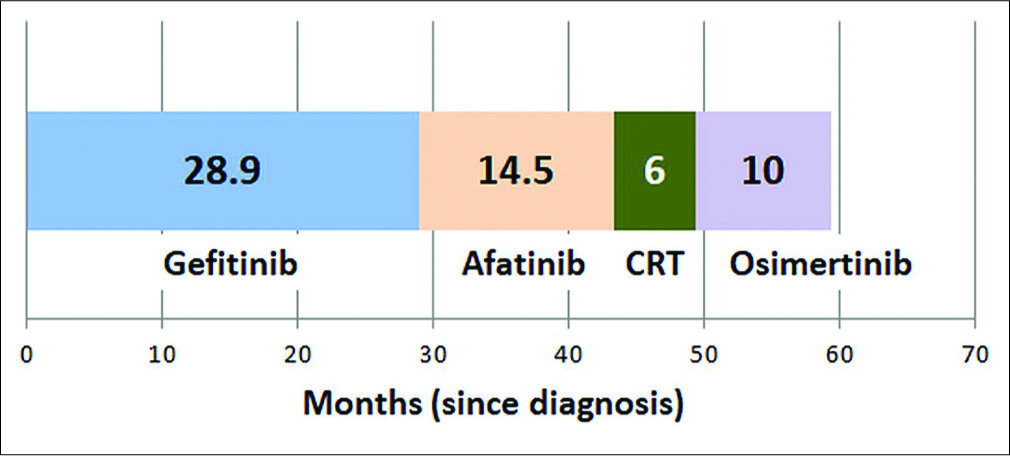
- Representative overall survival in patients with adenocarcinoma lung with epidermal growth factor receptor mutation (del19).
Such a situation is also applicable to other cancers, including breast cancer.[6,7] No wonder, the Indian Council for Medical Research guidelines on breast cancer clearly states that clinical utility of multigene signatures line MammaPrint and Oncotype Dx is uncertain because they are based on Caucasian population. Any test that has little data from the Indian patients is not ready for routine clinical use.[8] This is applicable irrespective from where the tissue or liquid biopsy is obtained from.[9]
To provide our patients with the benefit of such progress, we have to keep up to date with the evolving science and data. Moreover, medical oncologists seem to be ahead of the curve with more than half (63.5%) of all molecular testing being recommended by them, the rest coming from surgeons (15.3%), gynecologists (9.6%), patients themselves (6.2%), and general practitioners (2.6%).[10]
To disseminate this knowledge downstream, we have to understand the evolving terminology and the benefit of online resources.[11,12]
Let us take the example of hereditary breast cancer. We understand that risk is not straightforward Mendelian inheritance. While some germline variants disrupting gene function confer a high risk for cancer, many other variants (80–85%) have little or no clinical significance with respect to cancer risk. Earlier, we used to focus only on BrCa 1 and 2 genes. Now, we are wiser and have discovered several other high penetrance genes that confer risk of breast cancer (TP53, PTEN, PALB2, CHEK2, and ATM). Figure 2 shows the contribution of BrCa 1/2 genes versus others in three distinct ethnic groups. Table 1 shows the distribution of these mutations in a small cohort of these same ethnic groups studied in Singapore.[10]

- Ethnic comparison of pathogenic mutations (%) in patients with hereditary cancers.
| Mutation involving | Indian | Middle Eastern | Caucasian |
|---|---|---|---|
| BrCa1 | 5 | 3 | 9 |
| BrCa2 | 1 | 1 | 3 |
| TP53 and ATM | 3 | 0 | 0 |
| NEN and BR1P1 | 0 | 2 | 0 |
| CDH1 | 0 | 1 | 0 |
| Totalw | 9 | 7 | 12 |
When evaluating a risk of hereditary breast cancer, about 40% will have no mutation and the remaining will be equally divided amongst those having a known pathogenic mutation and those with a variation in the genetic sequence whose significance we are not sure of – the so-called variation of uncertain significance (VUS). Probands showing VUS might even have 2 (25%) or more (10%) such mutations. Moreover, the incidence of such VUS identification increases as better NGS methods are used or multigene panels become more extensive. Assigning the right certainty (either way – pathogenic or non-pathogenic) requires a careful correlation with clinical outcome IN THE SAME ETHNIC GROUP. Figure 3 shows the location of specific BrCa1 mutations amongst three ethnic groups, highlighting the differences.
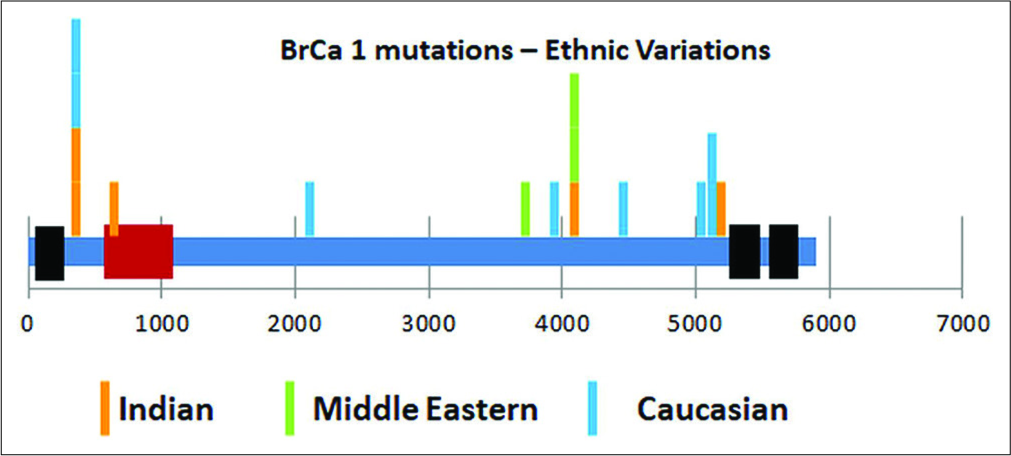
- Ethnic comparison of location of pathogenic mutations in BrCa1 gene.
The same is the case when we use molecular testing, protein expression, immunohistochemistry, or other biomarkers for assigning risk, commonly called as predictors of outcome.[13,14] We, therefore, strongly urge all academically inclined oncology colleagues to join hands in pooling data from Indian patients and help make our own data more robust, reduce the redundance of VUS, and help patients be assigned to the right personalized cancer management pathway.
Declaration of patient consent
Patient’s consent not required as there are no patients in this study.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- Gefitinib (IRESSA) in patients of Asian origin with refractory advanced non-small cell lung cancer: Subset analysis from the ISEL study. J Thorac Oncol. 2006;1:847-55.
- [CrossRef] [Google Scholar]
- Clinical experience with gefitinib in Indian patients. J Thorac Oncol. 2008;3:380-5.
- [CrossRef] [PubMed] [Google Scholar]
- Personalized medicine: Lung cancer leads the way. Ind J Cancer. 2013;50:77-9.
- [CrossRef] [PubMed] [Google Scholar]
- Patterns of epidermal growth factor receptor testing across 111 tertiary care centers in India: Result of a questionnaire-based survey. South Asian J Cancer. 2018;7:203-6.
- [CrossRef] [PubMed] [Google Scholar]
- Editorial-T790M mutation and clinical outcomes with genuine osimertinib. Indian J Med Paediatr Oncol. 2019;40:7-8.
- [CrossRef] [Google Scholar]
- Breast cancer: An overview of published Indian data. South Asian J Cancer. 2016;5:86-92.
- [CrossRef] [PubMed] [Google Scholar]
- Practical consensus recommendation on when to do BRCA testing. South Asian J Cancer. 2018;7:106-9.
- [CrossRef] [PubMed] [Google Scholar]
- ICMR Guidelines on Breast Cancer. Available from: https://www.icmr.nic.in/sites/default/files/guidelines/Breast_Cancer.pdf [Last accessed on 2020 Feb 20]
- [Google Scholar]
- Circulating tumor cells in breast cancer. Int J Mol Immunol Oncol. 2017;2:4-10.
- [CrossRef] [Google Scholar]
- Discoveries beyond BRCA1/2: Multigene testing in an Asian multi-ethnic cohort suspected of hereditary breast cancer syndrome in the real world. PLoS One. 2019;14:e0213746.
- [CrossRef] [PubMed] [Google Scholar]
- BRCA challenge: BRCA exchange as a global resource for variants in BRCA1 and BRCA2. PLoS Genet. 2018;14:e1007752.
- [CrossRef] [PubMed] [Google Scholar]
- Clinical implementation and validation of automated human genome variation society (HGVS) nomenclature system for next-generation sequencing-based assays for cancer. J Mol Diagn. 2018;20:628-34.
- [CrossRef] [PubMed] [Google Scholar]
- Analytical validation of CanAssist-breast: An immunohistochemistry based prognostic test for hormone receptor positive breast cancer patients. BMC Cancer. 2019;19:249.
- [CrossRef] [PubMed] [Google Scholar]
- Clinical validation of an immunohistochemistry-based CanAssist-breast test for distant recurrence prediction in hormone receptor-positive breast cancer patients. Cancer Med. 2019;4:1755-64.
- [CrossRef] [PubMed] [Google Scholar]





