Translate this page into:
Molecular profile of glioblastoma

-
Received: ,
Accepted: ,
How to cite this article: Biswas A, Morjaria SC. Molecular profile of glioblastoma. Int J Mol Immuno Oncol 2021;6(2):97-9.
Abstract
Glioblastoma multiforme (GBM) is one of the most devastating primary intracranial tumors in case of adults. The intricacy of GBM is contemplated at both the inter- and intra-tumoral levels by cellular and sub-molecular heterogeneity. Subsequently, various expected diagnostic, prognostic, and prescient biomarkers have been reflected. The current methodology and understanding of biomarkers of GBM are discussed here. We have also highlighted the molecular subtypes of GBM, genetic aberrations, and how the microRNAs are involved in glioblastoma.
Keywords
Astrocytic tumors
Biomarkers
Genetic aberrations
Tumor microenvironment
MicroRNAs
INTRODUCTION
Glioblastoma is a belligerent type of cancer that may occur in the brain or the spinal cord. It is also known as the glioblastoma multiforme (GBM). Glioma indicates the tumors that occur in the brain and the spinal cord. Glioblastoma usually forms from the astrocytes. Blastoma comes under the type of cancer which comes into effect due to the malignancies in the precursor cells generally known as the blast cells. Astrocytes are with star model cells with many processes which are the major and most several of the neuroglia. It provides strength to support neurons. According to the studies about 5% of all glioblastoma cases are hereditary and the other 95% do not chalk up to any specific cause.[1]
The glioblastoma cells possess more genetic anomalies as compared to the other forms of astrocytoma brain cancer. The genetic mutations that lead to glioblastoma progression can be the result of various causes including DNA defects that are inherited, frequent exposure to carcinogens or other chemicals or high dose exposure to ionizing radiation. Substantial immune cell infiltration is displayed by tumor microenvironment (TME) of human glioblastoma [Figure 1]. In the growth of glioblastoma, TME associated cell types are widely involved. Weighted Correlation Network Analysis (WGCNA) is a technique generally utilized for data mining, especially to scrutinize biological networks that are dependent on pairwise relationships between variables. WGCNA produced 14 notable modules, of which one contained genes which were active in extracellular matrix formation and immune response. The convergence of these genes with a GO immune-related gene set produced 47 immune related genes, of which five displayed more expression and participation with worse prognosis in glioblastoma.[2]
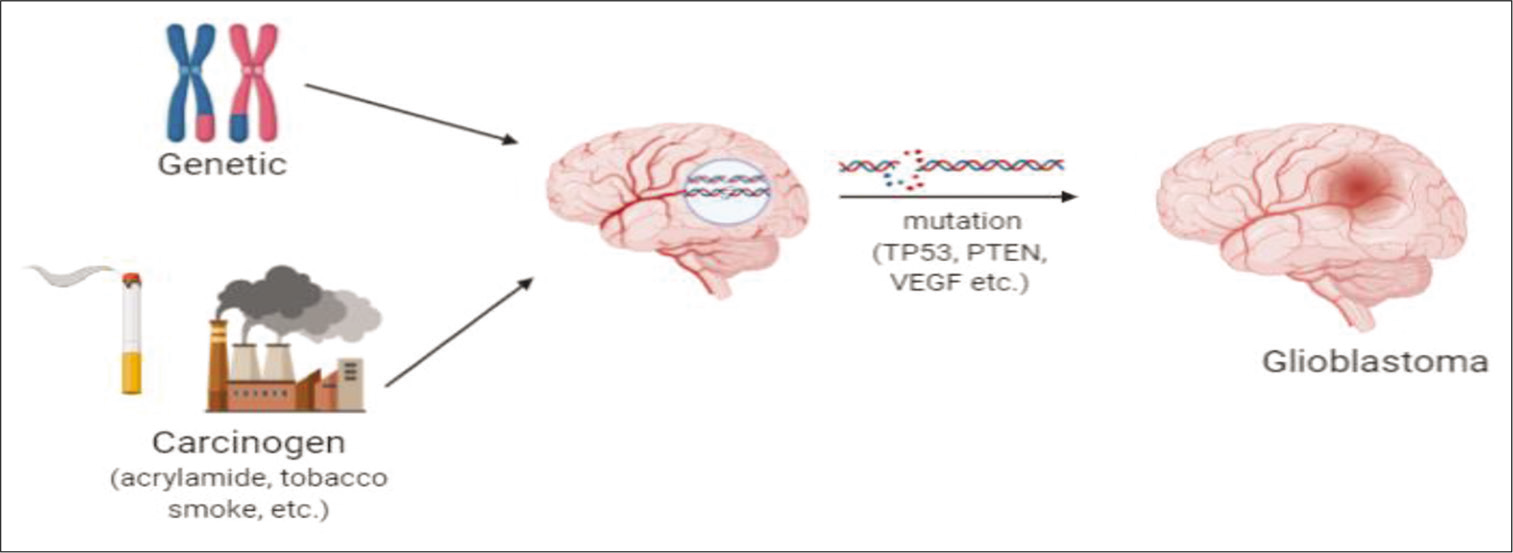
- Carcinogens undergoing mutation, resulting in the formation of glioblastoma.
GLIOBLASTOMA CLASSIFICATION
GBM can be categorized as primary (de novo), which is more generic and also very belligerent or as secondary glioblastoma which is not very common. GBM generally do make an appearance as Grade IV tumors and this is the most common form and older subjects are more prone to it. On the other hand, its secondary form generally may develop as lower grade astrocytic tumors (Grade II or III) later it may become more aggressive and evolve to Grade IV.[3]
MOLECULAR SUBTYPES
According to Phillips et al. proposal in 2006[4] of subtyping, gliomas have been classified, based on gene expression profiling as: Proneural, proliferative, and mesenchymal. The Cancer Genome Atlas (TCGA) network has, on the other hand, recognized four molecular subtypes of GBM, that is, proneural, neural, mesenchymal, and classical. GBM patients suffering from mesenchymal subtype possesses shorter life span as compared to the other subtypes.[5] Each of these subtypes is associated with specific genetic and epigenetic alterations.
BIOLOGICAL MARKERS
There are several biological markers still under assessment, whereas a few are currently accepted for diagnosis of GBM patients. These include isocitrate dehydrogenase, O6-methylguanine DNA methyltransferase (MGMT), epidermal growth factor receptor (EGFR), vascular endothelial growth factor, tumor suppressor protein (TP53), and phosphatase and tensin homolog. Others include phospholipid metabolites, p16INK4a gene, immature cancer stem cells, and imaging biomarkers.
At present, there are no clinically affirmed imaging biomarkers for GBM. However, advances in imaging strategies such as dynamic susceptibility-weighted contrast-enhanced perfusion imaging, dispersion weighted magnetic resonance imaging with apparent diffusion coefficient mapping, positron emission tomography, and magnetic resonance spectroscopy are being employed in studies aimed at classifying GBM tumors.[6]
MOLECULAR PATHWAYS AND GENETIC ABERRATIONS IN GLIOBLASTOMA
Tumor development in GBM cells is encouraged by high articulation of cell surface membrane receptors that control the intracellular signal transduction pathways controlling multiplication and cell cycle irregularities remembering an expansion for DNA repair proteins and abnormal cell death pathways. A coordinated examination of the hereditary modifications, performed by the TCGA research organization, indicated the role of pathways involving receptor tyrosine kinase through RAS/mitogen-enacted protein kinase and PI3K/AKT/mTOR, alongside cell cycle-regulating retinoblastoma tumor suppressor and p53.[6,7]
ROLE OF MICRORNAS IN GLIOBLASTOMA
MicroRNA (miRNA) comes under the group of non-coding RNA which is of about 22 bases in length. miRNAs bind to complementary sequences in the mRNA and reversibly modulate transcription through non-mutational mechanisms. The genes regulated by miRNAs in glioblastoma are associated with numerous pathways which include apoptosis, cell proliferation, autophagy, metastasis, and angiogenesis.[8] Since miRNAs have numerous targets in various tissues, they may have oncogenic or antioncogenic impact. The effects depend on the biological context, with oncogenic miRNAs also known as oncomiRs. For example, the various miRNAs that modulate the histone MGMT EZH2, on a basic level can be considered as tumor silencers. The miRNA let-7, hinders oncogenes such as MYC and K-RAS and consequently can hinder glioblastoma cell proliferation. Both miR-128 and miR-34a are examples of miRNAs that act as tumor suppressors. miR-128 is an antiproliferative miRNA that affects different pathways by influencing expression of EGFR, PDGFRA, WEE1, and E2F3a. miR-34a interferes with cell multiplication through various targets including CDK6, CCND1, and NOTCH.[8]
CONCLUSION
The molecular profile of glioblastomas is discussed. Further correlation of clinical observations with laboratory findings and molecular signatures is needed in the future. This will enable more refined classification of these tumors and unravel the mechanisms and potential individualized targets for treatment.
Acknowledgments
We thank Dr. Radhika Vaishnav for her mentorship, Mr. Arupam Biswas, Ms. Kavya Kadali, Ms. Rachna Gowlikar, and Ms. Syeda Nooreen for their helpful discussions during the Virtual Internship in Science Leadership conducted in 2020.
Declaration of patient consent
Patient’s consent not required as there are no patients in this study.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- Malignant gliomas. Curr Treatment Options Oncol. 2000;1:459-68.
- [CrossRef] [PubMed] [Google Scholar]
- Identification of immune-related genes contributing to the development of glioblastoma using weighted gene co-expression network analysis. Front Immunol. 2020;11:1281.
- [CrossRef] [PubMed] [Google Scholar]
- Aberrant signaling pathways in glioma. Cancers. 2011;3:3242-78.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157-73.
- [CrossRef] [PubMed] [Google Scholar]
- The landscape of the mesenchymal signature in brain tumours. Brain. 2019;142:847-66.
- [CrossRef] [PubMed] [Google Scholar]
- Diagnostic and therapeutic biomarkers in glioblastoma: Current Status and future perspectives. Biomed Res Int. 2017;2017:8013575.
- [CrossRef] [PubMed] [Google Scholar]
- The cancer genomic atlas “to conquer cancer. Int J Mol Immunol Oncol. 2020;2020:25259.
- [CrossRef] [Google Scholar]
- MicroRNA in glioblastoma: An overview. Noncoding RNAs in Health and Disease. 2017;2017:7639084.
- [CrossRef] [PubMed] [Google Scholar]






