Translate this page into:
Molecular determination of mutational signatures p53 and retinoblastoma (RB) in human papilloma virus-associated squamous cell carcinoma of the cervix

*Corresponding author: Emmanuel Akokhamen Omon, Department of Medical Laboratory Science, College of Medicine and Health Sciences, Afe Babalola University, AdoEkiti, Ekiti State, Nigeria. omonea@pg.abuad.edu.ng
-
Received: ,
Accepted: ,
How to cite this article: Ekundina VO, Omon EA. Molecular determination of mutational signatures p53 and retinoblastoma (RB) in human papilloma virus-associated squamous cell carcinoma of the cervix. Int J Mol Immuno Oncol 2024;9:53-61. doi: 10.25259/IJMIO_6_2024
Abstract
Objectives:
Cervical cancer is a cancer arising from the cervix due to the abnormal growth of cells that have the ability to invade or spread to other parts of the body, of which human papilloma virus (HPV) infection causes more than 90% of cases. This study aimed at investigating the mutational signatures retinoblastoma (RB) and p53 in HPV-associated squamous cell carcinoma (SCC) of the cervix.
Material and Methods:
A total of 10 formalin-fixed, paraffin-embedded tissue blocks, all consisting of SCC of the cervix, were used for this study. The technique employed was nucleic acid amplification technique and various steps for DNA sequencing including DNA extraction and polymerase chain reaction.
Results:
Mutation in the RB gene occurred at different gene regions ranging from 10 to 230, while p53 occurred at 10–945. The most prevalent mutational signature within the RB gene regions was T>C (37.50%), while the p53 gene region was T>A (23%). Percentage mutations at single-nucleotide polymorphisms (SNPs) of RB were transition (58.8%), transversion (41.2%), Indel (0%), and substitution (0%), while point mutations were missense (65%), silent (23%), and non-sense (12%), respectively. Mutations at SNPs of p53 were transversion (47.6%), transition (38.1%), Indel (14.3%), and substitution (0%), while point mutations were missense (78%), silent (22.2%), and non-sense (0%), respectively.
Conclusion:
Our results indicate that the inactivation of the normal functions of the tumor-suppressor proteins RB and p53 is an important step in human cervical carcinogenesis caused by mutation or from complex formation with the HPV oncoproteins.
Keywords
Mutational signatures
Cervical cancer
Human papilloma virus
p53
Retinoblastoma gene
INTRODUCTION
Human papilloma virus (HPV) is a non-enveloped, virus that carries double-stranded circular DNA. The virus lifecycle starts in the undifferentiated basal compartment of epithelia, where distal chromatin contacts between the early gene area and viral enhancer stabilize the viral chromatin, keeping it in an epigenetically repressed form.[1] Worldwide, HPV is associated with over 5% of all cancer cases. HPV is the most common sexually transmitted infection and one of the few viruses that can cause multiple benign or malignant cancers in more than 500,000 people each year.[2] Many malignancies, including those of the gastrointestinal tract, cervical, urinary bladder, and head and neck regions, are linked to HPV infection. These cancers are becoming a global concern and are responsible for the majority of cancer deaths in developing countries. HPVs are potentially associated with carcinogenesis in various categories of cancer.[3]
Cancer originating from the cervix is known as cervical cancer. The cause is attributed to atypical cellular proliferation that possesses the capacity to infiltrate or disperse throughout the body. Usually, no symptoms appear in the early stages. Subsequent symptoms could be pain during sex, pelvic pain, or irregular vaginal bleeding. Although bleeding after sexual activity might not be a severe concern, it could also be a sign of cervical cancer.[4] Cervical cancer is the fourth most frequent type of cancer worldwide and the fourth leading cause of cancer-related deaths among women. In low-income and middle-income countries, about 90% of deaths from cervical cancer occurred. Cervical cancer is the second most common cause of cancer-related deaths among women in Nigeria between the ages of 15 and 44 years, and it is also the third most common cancer overall.[5] Over 90% of cases are caused by an infection with the HPV, and the majority of people who have HPV infections do not go on to develop cervical cancer. About 70% and 25% of all cervix malignancies induced by HPV infection are squamous cell carcinoma (SCC) and adenocarcinoma, respectively.[6]
Retinoblastoma (RB) gene and tumor suppressor gene (p53) are well-known examples of cellular tumor suppressors that are involved in senescence, differentiation, apoptosis, cell cycle progression, DNA repair, and chromatin remodeling.[7] Crucially, p53 and/or RB functions are compromised in the great majority of human cancers. The gene p53, which is altered most frequently in human malignancies, is essential for controlling the growth and division of cells because it detects damage to DNA and triggers a biological reaction that either repairs or destroys the cell. The RB1 gene, which maps to chromosome 13q14 and encodes the tumor suppressor RB protein, is the source of RB due to mutational inactivation of both alleles.[8] About 60–70% of cervical cancer cases are histologically classified as cervical SCC. Cervical cancer is still the major cause of cancer-related deaths in women globally, even after screening and the introduction of the prophylactic HPV vaccine in developed nations.[9] Individuals with metastatic disease still have poor prognosis; therefore, there is need for novel treatments and effective molecular markers for patient stratification are urgently required. Thus, the purpose of this work was to determine the p53 and RB mutational signatures in HPV-associated SCC of the cervix.
MATERIAL AND METHODS
Ethics consideration
Ethical approval for this study was obtained from the Health Research Ethics Committee of Obafemi Awolowo University Teaching Hospital (OAUTH), IIe-Ife, Osun State, Nigeria (OAUTH/2023/0157). All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation and with the Helsinki Declaration of 1975, as revised in 2000.
Tissues sample collection
A total of ten formalin-fixed, paraffin-embedded tissue blocks, all consisting of SCC of the cervix, were retrieved from OAUTH, IIe-Ife, Osun State, for this case-control, retrospective investigation. This study was carried out between July 2023 and January 2024.
Molecular techniques
Molecular techniques for DNA sequencing were carried out under the following steps: DNA extraction, polymerase chain reaction (PCR), isolation of the gene of interest, integrity test, purification, and DNA sequencing.
DNA extraction
We extracted DNA from human tissue by following the Dellaporta DNA extraction methodology, with a few minor adjustments. In summary, 500 µL of extraction buffer was mixed with a sterile mortar and pestle to grind each tissue sample before it was placed in a sanitized Eppendorf tube. Following the addition of 33 µL of 20% sodium dodecyl sulfate, the mixture was vortexed and heated to 65°C for 10 min in an incubator. Following the incubation period, 10 µL of room temperature 5 M potassium acetate was added. This was followed by vortexing and centrifugation at 1000 × g for 10 min. After collecting the supernatant in a different Eppendorf tube, 300 µL of cold isopropanol was added, gently mixed and allowed to incubate for 60 min at 20°C. After centrifuging the DNA for 10 min at 13,000 × g, the pellet was carefully separated from the supernatant by gently decanting it without disturbing it. After centrifuging the DNA pellet at 1000 × g for 10 min, 500 µL of 70% ethanol was used as a wash. After the ethanol was decanted, the DNA was allowed to air dry at ambient temperature until the tube showed no signs of ethanol. To protect and suspend the DNA, the pellet was finally resuspended in 50 µL of Tris EDTA buffer.[10]
Polymerase Chain Reaction (PCR)
The PCR sequencing preparation cocktail for all PCRs was composed of 35 µL of sterile distilled water, 10 µL of ×5 GoTaq colorless reaction buffer, 3 µL of 25 mM MgCl2, 1 µL of 10 pmoL of each primer, and 0.3 µL of Taq DNA polymerase (Promega, USA). 1.5 µL of DNA template was also added. As shown in the Table 1, PCR was performed using a GeneAmp 9700 PCR System Thermocycler (Applied Biosystems Inc., USA) with a particular PCR profile for each primer.[11]
| Gene Name | Primer Name | Primer Sequence | PCR profile |
|---|---|---|---|
| TP53 | TP53F | GGGCGGATTACTTGAGGATAG | An initial denaturation at 94°C for 5 min; followed by a 30 cycle consisting of 94°C for 30s, 48°C for 30s and 72°C for 1 min and a final termination at 72°C for 10 min |
| TP53R | CCTAAACAGGACAAGGCAAATAC | An initial denaturation at 94°C for 5 min; followed by a 30 cycle consisting of 94°C for 30s, 48°C for 30s and 72°C for 1 min and a final termination at 72°C for 10 min |
PCR: Polymerase Chain Reaction
Integrity
The amplified gene fragment integrity was verified by electrophoresis on a 1.5% agarose gel. ×1 TAE buffer was used to create the gel, and 3µL of 0.5 g/mL ethidium bromide was used to stain it. Following gel solidification, 4 µL of each PCR product and 2 µL of ×10 blue gel loading dye were added to the gel. After 45 min of electrophoresis at 120 V, the gel was visible under ultra-violet transillumination. By comparing the mobility of the PCR products with a 100 bp DNA ladder run alongside the samples, the size of the PCR products was determined.[12]
Purification
Ethanol was used to purify the amplified fragments after gel electrophoresis. Subsequently, the separated pieces were examined on a 1.5% agarose gel and measured utilizing a Thermo Scientific NanoDrop Model 2000.[12]
Sequencing
Using the Big Dye Terminator v3.1 cycle sequencing kit and an Applied Biosystems Genetic Analyzer 3130 × l sequencer, the amplified fragments were sequenced. MEGA 6 and BioEdit tools were utilized for the genomic analysis.[13]
Data analysis
Data obtained from the study were analyzed in Microsoft Excel using simple frequency and percentage. Results were presented using tables, charts, and graphs, respectively.
RESULTS
Figure 1 depicts the Agarose gel showing the positive amplification of the RB gene amplified from DNA isolated from ten patients using RB gene-specific primers. The presence of 247bp indicates a positive amplification in extracted DNA. Figure 2 is an Agarose gel showing the positive amplification of the p53 (tumor suppressor) gene amplified from DNA isolated from 10 patients using p53 gene-specific primers. The presence of 900 bp indicates a positive amplification in extracted DNA. Figure 3 showed the visual representation of amino acid alignment with regions of functional mutation as a result of single nucleotide polymorphism (SNP) mutation along the p53 gene regions, while Figure 4 showed a visual representation of amino acid alignment resulting in the functional mutation of the RB gene region in SCC of the cervix. Figure 5 showed that SNPs in the RB gene often involve transitions (58.8%), while SNPs in the p53 gene frequently involve transversion (47.6%). Figure 6 showed that functional mutation in the RB and p53 gene often involved missense mutations (64.7% and 78%, respectively).
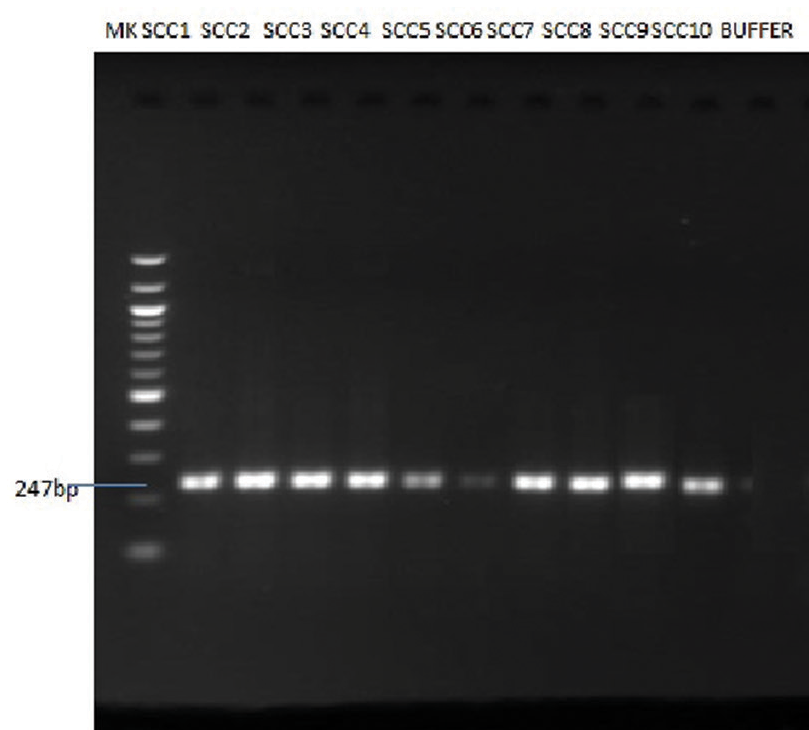
- Agarose gel showing the positive amplification of retinoblastoma (RB) gene amplified from DNA isolated from ten patients using RB gene-specific primers.
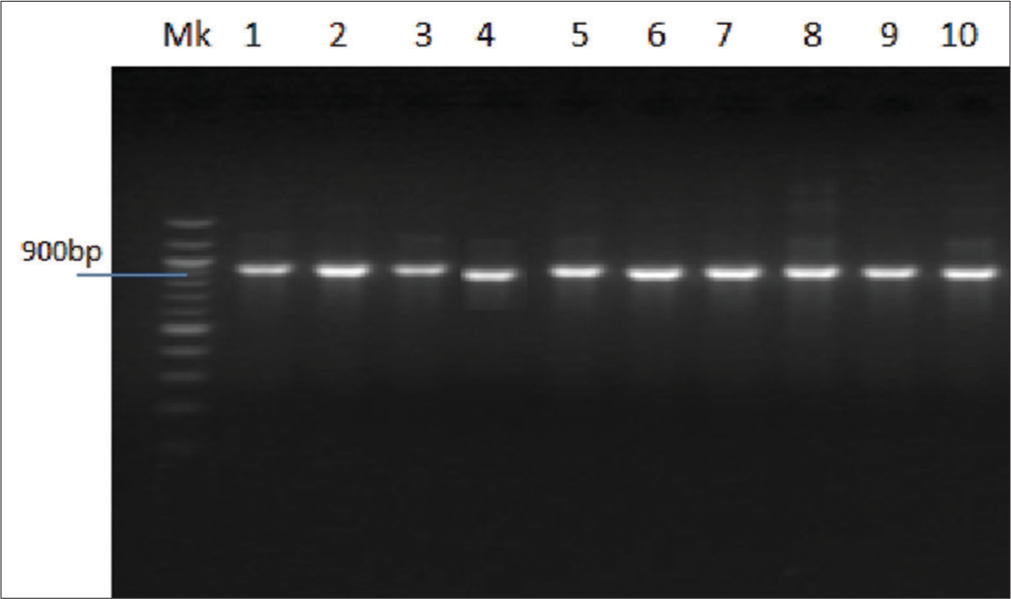
- Agarose gel showing the positive amplification of p53 (tumor suppressor) gene amplified from DNA isolated from five patients using p53 gene-specific primers.
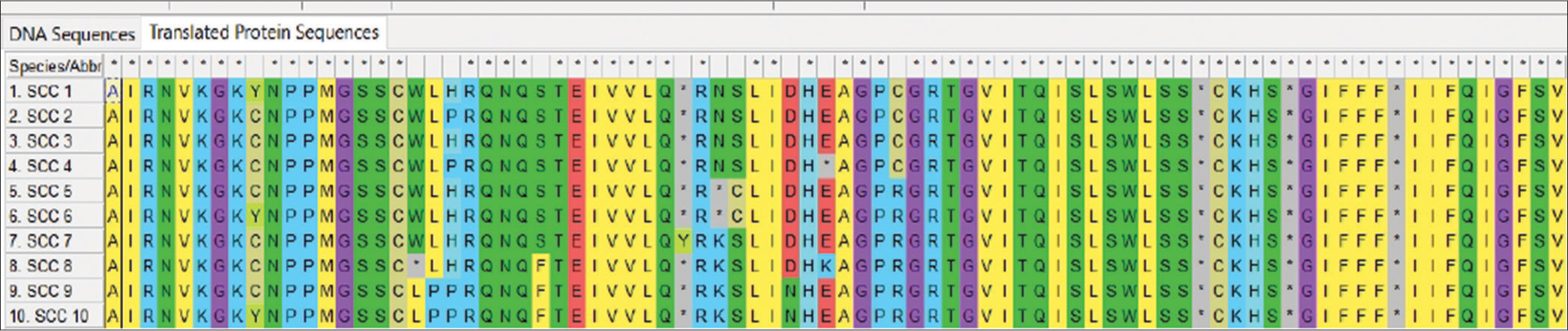
- Amino acid alignments showing regions of functional mutation as a result of single nucleotide polymorphism mutation along the retinoblastoma gene. SCC: Squamous cell carcinoma

- Amino acid alignments showing regions of functional mutations as a result of single nucleotide polymorphism mutation along the p53 gene regions. SCC: Squamous cell carcinoma
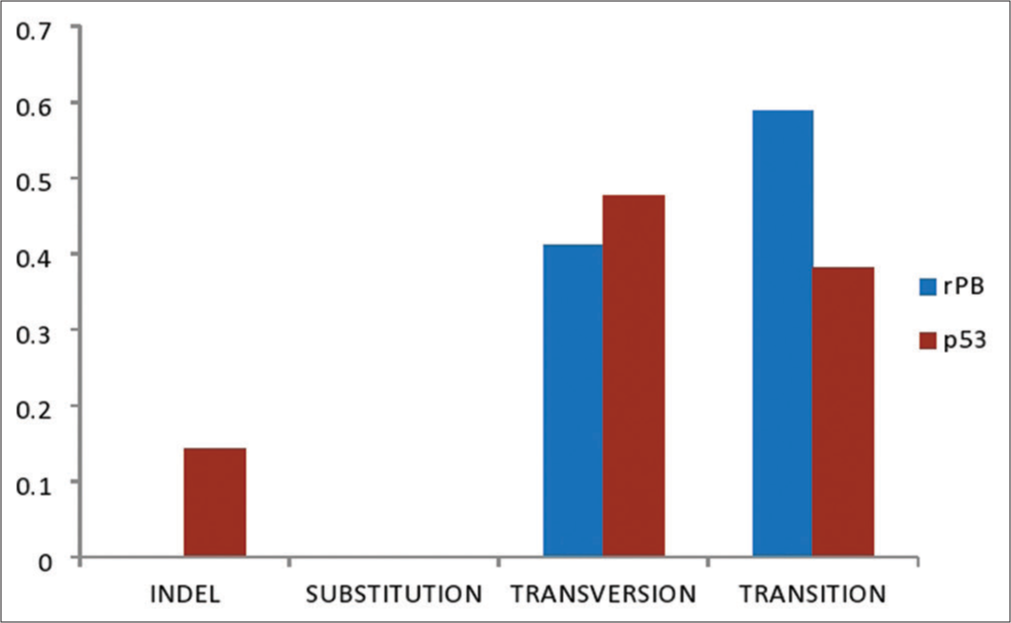
- A graph showing single nucleotide polymorphisms frequency in retinoblastoma and p53 genes of squamous cell carcinoma.
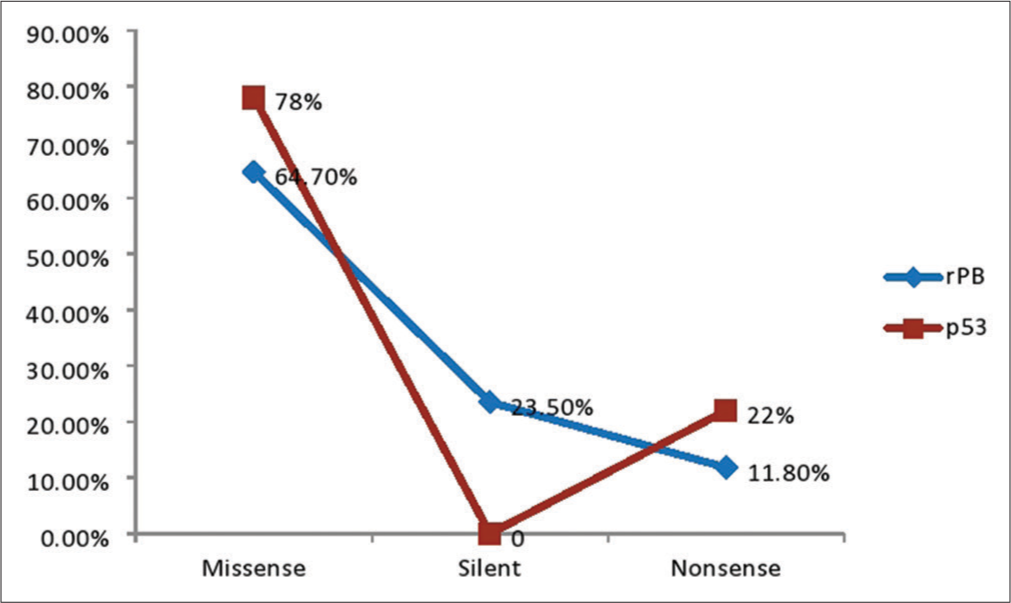
- A pie chart showing the frequency of the functional mutation type of retinoblastoma (pRb) and p3 genes in squamous cell carcinoma of the cervix.
Table 2 shows the clinical data of the subjects studied. Ten female subjects were recruited for this study, of which 20% belonged to the age group 20–30 years, 30% to the age group 31–40 years, and 50% belonged to the age group 41–50 years. The majority of the subjects were married (70%), Christians (70%), lived in urban areas (60%), had secondary education (40%), and were employed (40%).
| Variable | Frequency (%) |
|---|---|
| Age (years) | |
| 20–30 | 2 (20.0) |
| 31–40 | 3 (30.0) |
| 41–50 | 5 (50.0) |
| Marital status | |
| Single | 2 (20.0) |
| Married | 7 (70.0) |
| Divorced | 1 (10.0) |
| Educational status | |
| Primary | 3 (30.0) |
| Secondary | 4 (40.0) |
| Tertiary | 3 (30.0) |
| Occupation | |
| Government employed | 4 (40.0) |
| Self-employed | 4 (40.0) |
| Unemployed | 2 (20.0) |
| Religion | |
| Christian | 7 (70.0) |
| Muslim | 2 (20.0) |
| African traditional religion | 1 (10.0) |
| Residence | |
| Urban | 6 (60.0) |
| Rural | 4 (40.0) |
| Parity | |
| Nulliparous | 3 (30.0) |
| 1–2 births | 6 (60.0) |
| 3 and above | 1 (10.0) |
Table 3 showed that the most prevalent mutational signature within the RB gene regions is T>C (37.50%), followed by G>A (18.75%), T>G>A (12.50%) and the least prevalent is the C>A (6.25%), C>G>A (6.25%), and T>G (6.25%), respectively, while Table 4 showed the gene mutation, the location, the mutation type, and their descriptions within fragments of the RB gene in regions ranging from 10 to 230 in squamous cell carcinoma of the cervix. Table 5 showed that the most prevalent mutational signature within the p53 gene regions is T>A (23%), followed by C>T (19.0%), A>G (19.0%), C>G (19.0%), and T (9.5%), while T>G (5%) and C (5%) were the least prevalent, while Table 6 showed the gene mutation, the location, the mutation type, and their descriptions within fragments of the tumor suppressor P58 gene in regions ranging from 25 to 945 in squamous cell carcinoma of the cervix.
| Mutational Signature | Number of occurrences (Frequency) | Percentage |
|---|---|---|
| C>A | 1 | 6.25 |
| T>A | 2 | 12.50 |
| T>G | 1 | 6.25 |
| G>A | 3 | 18.75 |
| T>C | 6 | 37.50 |
| C>G > A | 1 | 6.25 |
| T>G > A | 2 | 12.50 |
RB: Retinoblastoma
| Description | Location | Gene type | Specimen | Mutation type | Mutation type description |
|---|---|---|---|---|---|
| C>A | 59 (7:3) | RB | SCC | Transversion | Missense mutation changes histidine to proline |
| T>A | 103 (1:4) | RB | SCC | Transversion | Missense mutation changes stop code to lysine |
| T>A | 106 (1:4) | RB | SCC | Transversion | Missense mutation changes cysteine to serine |
| T>G | 105 (3:2) | RB | SCC | Transversion | Missense mutation changes asparagine to lysine |
| G>A | 26 (7:3) | RB | SCC | Transition | Missense mutation changes cysteine to tyrosine |
| G>A | 93 (7:3) | RB | SCC | Transition | Silent mutation as amino acid remained leucine |
| G>A | 115 (4:1) | RB | SCC | Transition | Missense mutation changes aspartate to asparagine |
| T>C | 56 (8:2) | RB | SCC | Transition | Missense mutation changes leucine to proline |
| T>C | 41 (9:1) | RB | SCC | Transition | Silent mutation as amino acid remained glycine |
| T>C | 69 (4:1) | RB | SCC | Transition | Silent mutation as amino acid remained asparagine |
| T>C | 74 (7:3) | RB | SCC | Transition | Missense mutation changes serine to phenylalanine |
| T>C | 84 (7:3) | RB | SCC | Transition | Silent mutation as amino acid remained isoleucine |
| T>C | 133 (3:2) | RB | SCC | Transition | Missense mutation changes cysteine to arginine |
| C>G > A | 99 (7:1:3) | RB | SCC | Transversion | Change stop code to tyrosine |
| T>A > G | 53 (7:1:2) | RB | SCC | Transition/Transversion | Nonsenses mutation and missense mutation changes tryptophan to a stop code and leucine respectively |
| T>A > G | 53 (8:1:1) | RB | SCC | Transition/Transversion | Nonsense mutation and missense mutation changes glutamic acid to a stop code and lysine, respectively |
RB: Retinoblastoma, SCC: Squamous cell carcinoma
| Mutational signature | Number of occurrence | Percentage |
|---|---|---|
| C | 1 | 5 |
| T | 2 | 9.5 |
| C>G | 4 | 19.0 |
| A>G | 4 | 19.0 |
| T>G | 1 | 5 |
| C>T | 4 | 19.0 |
| T>A | 5 | 23 |
| Description | Location | Gene type | Specimen | Mutation Type | Mutation type description |
|---|---|---|---|---|---|
| C | 231 (1:4) | P53 | SCC | Indel | Insertion of asparagine |
| T | 798 (1:1) | P53 | SCC | Indel | Insertion of leucine |
| T | 850 (1:1) | P53 | SCC | Indel | Insertion of phenylalanine |
| C>G | 25 (3:7) | P53 | SCC | Transversion | Missense mutation changing glutamine to glutamic |
| C>G | 35 (7:3) | P53 | SCC | Transversion | Missense mutation changing valine to leucine |
| C>G | 118 (7:3) | P53 | SCC | Transversion | Missense mutation changing glycine to arginine |
| C>G | 124 (7:3) | P53 | SCC | Transversion | Missense mutation changing valine to leucine |
| T>A | 51 (7:3) | P53 | SCC | Transversion | Missense mutation changing asparagine to lysine |
| T>A | 831 (1:1) | P53 | SCC | Transversion | Missense mutation changing histidine to leucine |
| T>A | 863 (1:1) | P53 | SCC | Transversion | Missense mutation changing cysteine to serine |
| T>A | 925 (3:2) | P53 | SCC | Transversion | Missense mutation changing serine to arginine |
| T>A | 945 (1:1) | P53 | SCC | Transversion | Missense mutation changing stop code to leucine |
| T>G | P53 | SCC | Transversion | Silent mutation retaining leucine | |
| C>T | 84 (3:7) | P53 | SCC | Transition | Silent mutation retaining alanine |
| C>T | 117 (3:5) | P53 | SCC | Transition | Missense mutation changing phenylalanine to leucine |
| C>T | 149 (7:3) | P53 | SCC | Transition | Missense mutation changing proline to leucine |
| C>T | 181 (4:1) | P53 | SCC | Transition | Missense mutation changing cysteine to arginine |
| A>G | 188 (3:2) | P53 | SCC | Transition | Missense mutation changing tyrosine to cysteine |
| A>G | 204 (2:3) | P53 | SCC | Transition | Silent mutation retaining alanine |
| A>G | 211 (7:3) | P53 | SCC | Transition | Missense mutation changing threonine to alanine |
| A>G | 213 (3:7) | P53 | SCC | Transition | Silent mutation retaining threonine |
SCC: Squamous cell carcinoma
DISCUSSION
In this study, the mutation in the p53 tumor suppressor gene showed occurrence at different gene regions ranging from 10 to 945. This finding is in agreement with the previous studies.[14-16] The complex formation between the viral oncoproteins and p53 is thought to inactivate the normal function of p53 in regulating cell proliferation.[15] p53 mutations fall into two primary categories: Structural and DNA-contact mutations. These mutations impact the folding of the p53 protein or the transcriptional activity of p53, which, in turn, controls the expression of target genes. In both situations, the structural stability of p53 is changed, and the protein may develop a gain- or loss-of-function phenotype.[17] Changes in gene expression are one of the main effects of p53 mutant activities [18], and most of the time, this results in a significant reduction in the mutant’s capacity to bind canonical p53 elements. Tumorigenesis frequently results in the p53 tumor suppressor becoming inactive. The p53 gene is mutated in the majority of cases, resulting in a stable mutant protein whose buildup is thought to identify cancer cells. Mutant p53 proteins frequently acquire new oncogenic properties that give cells growth and survival advantages in addition to losing their tumor-suppressive properties.[19] It is noteworthy that the p53 gene mutations seen in this study happened at distinct stages of the multistep process of HPV-associated cervix SCC, hence differing in their contributions to the aggressiveness, metastasis, and genesis of the tumor. As a result, p53 gene mutation molecular analysis may be helpful as a cervical cancer screening biomarker.
RB gene is a regulatory protein in cell cycle proliferation. The RB mutation in this study happened at several gene locations, spanning from 10 to 230. This result is consistent with other research.[20-22] Mutations in RB have been discovered to correspond to areas of the cellular protein involved in complexing with the viral oncoproteins in human RBs and other malignancies, including cervical cancers.[20] It is possible that the mutant forms of RB in cervical cancer cells are also less able to bind to the normal cellular targets of RB because they are unable to form complexes with viral oncoproteins.[21] Furthermore, the phosphorylation capacity of these RB mutants is compromised. Mutations in RB have been discovered to correspond to areas of the cellular protein involved in complexing with the viral oncoproteins in human RBs and other malignancies, including cervical cancers.[20] It is possible that the mutant forms of RB in cervical cancer cells are also less able to bind to the normal cellular targets of RB because they are unable to form complexes with viral oncoproteins.[21] Furthermore, the phosphorylation capacity of these RB mutants is compromised.[22] These changes could be the result of HPV infection, which modifies the amino acid sequence and interferes with the capacity of RB to engage with its binding partners, making it difficult for RB to properly control cell division.[23]
In this study, mutational signatures in each gene were identified using the nucleic acid amplification technique. The most common mutation in the RB gene was T>C, indicating that cytosine (C) comes before thymine (T) as a result of the molecular mechanism linked to HPV infection. The E7 oncoprotein causes genetic changes, such as certain mutations in the DNA sequence, and damages DNA by specifically targeting the RB gene. The observed T>C mutations are caused by these mutations, which frequently entail the conversion of T nucleotides to C bases.[24] This finding is in agreement with the previous studies.[23,24] The apolipoprotein B mRNA editing catalytic polypeptide-like (APOBEC) family of cytidine deaminases is responsible for T>C mutations. In HPV-infected cells, this enzyme’s activity is increased, which results in a distinctive mutation pattern that is frequently observed in HPV-mediated malignancies like cervical cancer.[25] The conversion of cytidine to uridine by APOBEC enzymes can result in base pair formation with adenosine, which can lead to T>C substitution mutations following replication. The genomes of patients with cervical cancer have been found to include APOBEC-related alterations, and because the HPV genome plays a part in the host defense against viral infections, it is also prone to APOBEC editing.[26,27]
On the other hand, the most common mutational signature of the tumor suppressor gene (p53) is T>A, indicating that thymine (T) comes before adenine (A). This is linked to the molecular process of HPV infection, which is in line with other research findings.[13,27,28] These mutations lead to a changed p53 protein, which is unable to properly control cell division or induce apoptosis in cells that have damaged or mutant DNA. DNA damage can, therefore, build up within cells. Tumors may develop from these cells if they continue to divide uncontrollably.[27] The normal activity of p53 and other tumor suppressor genes is disrupted by the viral oncoprotein E6 generated by high-risk HPV strains. By attaching itself to the cellular p53 protein and encouraging its destruction, the E6 oncoprotein stops the protein from performing its typical tumor suppressor functions.[28] Cervical cancer is among the many human cancers that have been linked to the initiation and spread of p53 gene dysfunction and inactivation. It is well known that HPV virus oncoproteins control host tumor suppressor proteins like p53, which result in the malfunctioning of tumor suppressor protein.[16]
On the SNPs in RB, transition mutation was the most prevalent mutation (58.8%), followed by transversion (41.2%), while Indel and substitution mutation were both absent (0%). This finding shows that transition mutation has an increased prevalence in the RB gene compared to any other type of mutation, which is in agreement with the previous study.[29-31] Mutations in conserved areas of the RB and p53 genes are present in cervical tumors that overexpress the p53 protein, just like in other human cancer types. Since most of these mutations are transitions, it seems more likely that they developed naturally than being caused by exposure to carcinogens.[31] On the other hand, for tumor suppressor gene p53, transversion (47.6%) was the most prevalent mutation, followed by transition (38.1%), Indel (14.3%), and substitution (0%), respectively. The results align with earlier research, indicating that transversion mutations in the p53 gene are more common.[32] This finding has multiple contributing factors. Occasionally, lesions can result in additional nucleotides being introduced into the backbone or bases being skipped during replication. In addition, when sister chromatids and/or homologous chromosomes are separated from one another during the processes of mitosis and meiosis, mutations may also develop.[31]
In the SCC of the cervix, the frequency of the point mutation of the RB genes was as follows: missense mutation (65%), silent (23%), and nonsense (12%), respectively. On the other hand, the sequence analysis of p53 gene fragments isolated by polymerase chain reaction showed that missense mutation had the highest frequency (78%), followed by silent mutation (22%) and non-sense mutation (0%), respectively. Although a clear connection between silent p53 gene alterations and a poor prognosis for breast cancer has been reported,[33] there is currently no proof of this for cervical cancer. It is possible that missense and silent mutations work in concert to cause cancer.[33] Missense mutations in the p53 gene cause single amino acid substitutions, which impair the p53 protein structure and render it inactive.[18] Consistent with earlier reports, missense mutations were the most frequently found mutations in our analysis.[17,33] The majority of the mutations found in this study were missense and silent mutations, which are consistent with other findings suggesting that the RB gene and the tumor suppressor gene (p53) do not accrue mutations readily or spontaneously in cervical carcinogenesis.[34] As a result, a mutation in this gene may be a highly significant biomarker for assessing the onset and rate of advancement of cervical cancer.
CONCLUSION
The study concludes that RB and p53 regulatory functions are commonly annulled in human cervical cancers, either by mutation or as a consequence of their complex formation with the HPV oncoproteins. This study provides evidence that p53 and RB are relevant targets in cervical carcinogenesis. Therefore, understanding these mutational signatures is crucial for improving knowledge of the underlying mechanisms of HPV-related cervical cancer aid in the development of targeted therapies (gene therapies), early detection methods, and personalized treatment approaches for individuals affected by this disease.
Authors’ contributions
VOE: Conceptualization and design. VOE and EAO: Data acquisition and data analysis. EAO: Statistical analysis. EAO: Manuscript preparation. VOE, EAO: Manuscript editing and review. VOE and EAO: Approval of final manuscript.
Ethical approval
The Research/Study is approved by the Health Research Ethics Committee of Obafemi Awolowo University Teaching Hospital (OAUTH), IIe-Ife, Osun State, Nigeria, Number (OAUTH/2023/0157).
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent.
Conflicts of interest
There are no conflicts of interest.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
Financial support and sponsorship
Nil.
References
- Epigenetic regulation of human papillomavirus transcription in the productive virus life cycle. Semin Immunopathol. 2020;42:159-71.
- [CrossRef] [Google Scholar]
- HPV16 and HPV18 genome structure, expression, and post-transcriptional regulation. Int J Mol Sci. 2022;23:4943.
- [CrossRef] [Google Scholar]
- Protein-DNA interactions regulate human papillomavirus DNA replication, transcription, and oncogenesis. Int J Mol Sci. 2023;24:8493.
- [CrossRef] [Google Scholar]
- The complexity of human papilloma virus in cancers: A narrative review. Infect Agent Cancer. 2023;18:13.
- [CrossRef] [Google Scholar]
- Addressing missed opportunities for cervical cancer screening in Nigeria: A nursing workforce approach. Ecancermedicalscience. 2022;16:1373.
- [CrossRef] [Google Scholar]
- Cervical cancer, different treatments and importance of bile acids as therapeutic agents in this disease. Front Pharmacol. 2019;10:484.
- [CrossRef] [Google Scholar]
- Cell cycle regulation: P53-p21-RB signaling. Cell Death Differ. 2022;29:946-60.
- [CrossRef] [Google Scholar]
- Genomic analysis of BAX and Bcl-2 gene mutations in human papilloma virus-associated squamous cell carcinoma of the cervix. Surg Exp Pathol. 2024;7:11.
- [CrossRef] [Google Scholar]
- Integrated analysis of cervical squamous cell carcinoma cohorts from three continents reveals conserved subtypes of prognostic significance. Nat Commun. 2022;13:5818.
- [CrossRef] [Google Scholar]
- The study of mutations of CDK4 and TP53 genes in selected breast lesions. Sokoto J Med Lab Sci. 2023;8:65-77.
- [CrossRef] [Google Scholar]
- The study of BRCA1 and BRCA2 gene mutations in benign and malignant lesions of the breast. Sokoto J Med Lab Sci. 2023;8:122-32.
- [CrossRef] [Google Scholar]
- The Predictive significance of P16INK4A, Ki-67 and Ck7 expressions in the progression to SCC of the cervix. Int J Sci Eng Technol. 2021;8:195-208.
- [Google Scholar]
- Human papillomavirus E6 and E7: The cervical cancer hallmarks and targets for therapy. Front Microbiol. 2019;10:3116.
- [CrossRef] [Google Scholar]
- Assessment of P53 BCL-2 and CD34 in premalignant and malignant cervical lesions. J Krishna Inst Med Sci Univ. 2022;11:31-45.
- [Google Scholar]
- Metabolic functions of the tumor suppressor p53: Implications in normal physiology, metabolic disorders, and cancer. Mol Metab. 2020;33:2-22.
- [CrossRef] [Google Scholar]
- Exploring the p53 connection of cervical cancer pathogenesis involving north-east Indian patients. PLoS One. 2020;15:e0238500.
- [CrossRef] [Google Scholar]
- Mutational analysis of p53 gene in cervical cancer and useful polymorphic variants in exons 3 and 4. Egypt J Med Hum Genet. 2021;22:23.
- [CrossRef] [Google Scholar]
- Assessment of CD34, PSMA and p53 IHC expression in normal, benign and malignant prostate lesions. Bayero J Med Lab Sci. 2022;7:1-15.
- [Google Scholar]
- Targeting mutant p53 for cancer therapy: Direct and indirect strategies. J Hematol Oncol. 2021;14:157.
- [CrossRef] [Google Scholar]
- Retinoblastoma tumor suppressor protein roles in epigenetic regulation. Cancers (Basel). 2020;12:2807.
- [CrossRef] [Google Scholar]
- Loss of tumor suppressor gene function in human cancer: An overview. Cell Physiol Biochem. 2018;51:2647-93.
- [CrossRef] [Google Scholar]
- Retinoblastoma protein paralogs and tumor suppression. Front Genet. 2022;13:818719.
- [CrossRef] [Google Scholar]
- Molecular mechanisms in progression of HPV-associated cervical carcinogenesis. J Biomed Sci. 2019;26:28.
- [CrossRef] [Google Scholar]
- The drivers, mechanisms, and consequences of genome instability in HPV-driven cancers. Cancers (Basel). 2022;14:4623.
- [CrossRef] [Google Scholar]
- APOBEC: A molecular driver in cervical cancer pathogenesis. Cancer Lett. 2021;496:104-16.
- [CrossRef] [Google Scholar]
- Mutagenic activity of AID/APOBEC deaminases in antiviral defense and carcinogenesis. Mol Biol. 2022;56:46-58.
- [CrossRef] [Google Scholar]
- Ubiquitination of the HPV oncoprotein E6 is critical for E6/E6AP-mediated p53 degradation. Front Microbiol. 2019;10:2483.
- [CrossRef] [Google Scholar]
- Human oncoviruses and p53 tumor suppressor pathway deregulation at the origin of human cancers. Cancers (Basel). 2018;10:213.
- [CrossRef] [Google Scholar]
- Transition-transversion encoding and genetic relationship metric in relief feature selection improves pathway enrichment in GWAS. BioData Min. 2018;11:23.
- [CrossRef] [Google Scholar]
- Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim Biophys Acta Rev Cancer. 2021;1876:188556.
- [CrossRef] [Google Scholar]
- The mechanism of replication stalling and recovery within repetitive DNA. Nat Commun. 2022;13:3953.
- [CrossRef] [Google Scholar]
- Recent findings on the role of wild-type and mutant p53 in cancer development and therapy. Front Mol Biosci. 2022;9:903075.
- [CrossRef] [Google Scholar]
- P53 and Ki67 expression by cervical cancers in Ile-Ife, Nigeria. Br J Med Med Res. 2016;17:1-9.
- [CrossRef] [Google Scholar]
- TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2:a001008.
- [CrossRef] [Google Scholar]







