Translate this page into:
Multiple gene silencing in STAT pathway in K562 cells

*Sudha Shrikant Deo, Department of Immunology, Sir H N Medical Research Society, Mumbai, Maharashtra, India. Sudha.deo@rfhospital.org
-
Received: ,
Accepted: ,
Abstract
Context:
Chronic myeloid leukemia (CML) is characterized by the presence of a fusion oncoprotein BCR-ABL. This mutation imparts a constitutive phosphorylation activity of tyrosine residues in the cellular proteins. One of the targets of BCR-ABL is the STAT5 protein, which when phosphorylated induces gene expression of antiapoptotic proteins such as BCL-XL. The STAT pathway has been targeted in the past by disrupting any one protein only. A multiple gene silencing has never been done in this pathway.
Aim:
The aim of this study was to compare the effects of downregulation of BCR-ABL, STAT5A, STAT5B, and BCL-XL, individually and simultaneously, in human CML cell line (K562 cells) through RNA interference (RNAi). Further, gene expression, inhibition of proliferation, and apoptosis induction were assessed in K562 cells.
Materials and Methods:
K562 cells were transfected with various combinations of small iRNA (siRNA) and the expressions of aforesaid genes were determined by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blot analysis. K562 cell proliferation and apoptosis were analyzed using 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide and flow cytometry, respectively. The results were compared through one-way analysis of variance.
Results:
qPCR and western blotting results post-siRNA transfection confirmed the targeted gene suppression and protein reduction in K562 cells. The cell proliferation assay and apoptosis assay revealed that simultaneous gene silencing of BCR-ABL, STAT5A, STAT5B, and BCL-XL had the highest killing effect on K562 cells as compared to knocking down these genes individually or in any other combinations.
Conclusions:
This was the first time it was shown that multiple gene silencing in STAT pathway in CML cell line K562 was better as compared to individual gene silencing.
Keywords
BCL-XL
BCR-ABL
K562
Small interference RNA
STAT5
INTRODUCTION
It is estimated that there will be 23.6 million new cases of cancer each year by 2030. Chronic myeloid leukemia (CML) itself has an incidence of 1–2 cases per 100,000 adults and it accounts for 30–60% of all adult leukemia in India.[1] CML is identified by a reciprocal translocation t(9;22) (q34;q11), known as Philadelphia (Ph) chromosome. This results in the formation of a unique gene product (P210) displaying a constitutive tyrosine kinase activity. The deregulated tyrosine kinase transforms healthy cells into cancerous and has become a primary target for the treatment of CML.[2] The Ph chromosome has been observed to be present in the bone marrow in 95% of cases of CML.[3]
The standard first line of treatment for patients in chronic phase of CML is tyrosine kinase inhibitor (TKI), imatinib mesylate (trade name: Gleevec). Imatinib mesylate selectively induces the growth arrest and the apoptosis of BCR-ABL-positive leukemia cells.[4] Though Imatinib was widely used for CML, a mutation known as T315I, where threonine was substituted with isoleucine, rendered the drug no longer effective to bind to the ATP domain in BCR-ABL oncoprotein.[5] Except for a third-generation TKI (ponatinib), all the other TKIs (dasatinib, nilotinib, and bosutinib) are not effective against T315I mutation in the kinase domain of BCR-ABL oncoprotein.[6,7] Moreover, mutations might occur in various genes of a pathway with variable frequencies. Along with targeting various key regulators in cancer, it would be a better approach to target an entire pathway in any cancer.[8]
BCR-ABL fusion gene plays the central role in CML and has been shown to participate in many pathways such as MAPK, RAS, RAF, JUN kinase, and MYC.[9] One of the important pathways in CML is the STAT pathway which includes STAT5 gene as a downstream component of the JAK/STAT pathway. BCR-ABL directly instigates the tyrosine phosphorylation and dimerization of STAT5, followed by translocation of STAT5 dimers to the nucleus, wherein they activate many antiapoptotic genes.[10] It has also been reported that BCL-XL is induced by BCR-ABL through the activation of STAT5 in cell lines from CML patients.[11]
Small interference RNA (iRNA) therapy is speedily finding its way to get established as a dependable line of treatment.[12] Several researchers have tried to silence the CML-associated genes individually at the mRNA level such as BCR-ABL,[13] STAT5,[14] BCL-XL,[15] and many other genes.[16] Some researchers have tried silencing two genes in CML[17] and others have tried silencing multiple genes in various disorders such as breast cancer.[18]
Therefore, in this study, we hypothesized that the simultaneous gene silencing of the key oncogene (BCR-ABL) in CML and additional STAT5 pathway disruption might be highly effective in killing K562 cells (human CML cell line). For this purpose, siRNA against BCR-ABL, STAT5A, STAT5B, and BCL-XL was transfected in K562 in all the 15 possible gene combinations. The effect of downregulating BCR-ABL, STAT5A, STAT5B, and BCL-XL gene expressions was analyzed by studying the proliferation inhibition and apoptosis induction in K562 cells. Comparing the results obtained, it was concluded that silencing all four genes together induced a higher rate of apoptosis in K562 cells.
MATERIALS AND METHODS
Cell Culturing
K562 cells were obtained from the National Center for Cell Science (Pune, India). The cells were cultured in Iscove’s Modified Dulbecco’s Medium (IMDM, Gibco, USA) supplemented with 10% fetal bovine serum (Gibco, USA) and antibiotics penicillin (100 kU/L) and streptomycin (100 mg/L) at 37°C in a humidified incubator with 5% CO2.
Gene Knockdown in K562
ON-TARGETplus siRNAs for BCR-ABL, STAT5A, STAT5B, and BCL-XL were purchased from Dharmacon, USA. BCR-ABL siRNA was synthesized using the following sequence: 5'GCAGAGUUCAAAAGCCCUUdTdT 3,[13] and the remaining siRNAs were ordered off the shelf. From our previous experiment, it was decided to use 50 nM of each siRNA for 48 h for every experiment unless specified otherwise. However, to ensure comparability among experiments, a total amount of 200 nM siRNA were always used. For example, when only one target siRNA (50 nM) was used, then the remaining 150 nM concentration was filled by non-targeting (NT) siRNA to attain a total concentration of 200 nM. Similarly, when two siRNAs (50 nM + 50 nM) targeting two different genes were used, then 100 nM of NT siRNA was added to make the final concentration of 200 nM. Moreover, finally, when three siRNAs (150 nM) targeting three different genes were used, then 50 nM of NT siRNA was added to the mixture. No NT siRNA was added when all four siRNAs were used together as the total concentration of the siRNA mixture was already 200 nM. Although in the text, only the concentration of target siRNA used would be mentioned. The control cells were always transfected using 200 nM of NT siRNA for 48 h in all the experiments performed.
All of the transfections were conducted using a HiPerFect Transfection Kit (Qiagen, USA) according to the manufacturer’s instructions. Briefly, 2 × 105 K562 cells were plated per well of a 24-well plate in 100 μl serum containing IMDM. The required amount of siRNA was diluted in 100 μl serum-free IMDM, and 6 μl of HiPerFect Transfection Reagent was dissolved to the diluted siRNA for 5 min at room temperature. The complex (106 µl) was added to the cells, gently shaken, and thoroughly incorporated. After 6 h, 400 μl culture medium containing serum was added to the cells and incubated until further analysis. siRNA transfection efficiency was analyzed by counting positively transfected cells by SiGLO Red siRNA (Dharmacon, USA) per 100 cells under fluorescent microscope. The cells transfected with only Transfection Reagent HiPerFect were used as reference sample and cells transfected with NT siRNA (Dharmacon, USA) were used as control for all experiments.
Quantitative Polymerase Chain Reaction (Qpcr)
Total RNA was extracted from the sample by NucleoSpin® RNA/Protein kit by Macherey-Nagel GmbH & Co. (Germany) and reversed transcribed into cDNA by utilizing High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA) according to the manufacturer’s instructions. The primer sequences were as follows:
BCR-ABL forward, 5’- GTGTGAAACTCCAGACTGTC -3’, and reverse, 5’- CAAAATCATACAGTGCAACGA -3’,[19]
STAT5A forward, 5’- GAAGCTGAACGTGCAC-ATGAATC -3’, and reverse, 5’- GTAGGGACAGAGTCTTCACCTGG -3’,[14]
STAT5B forward, 5’- AGTTTGATTCTCAGGAAAGAATGT -3’, and reverse, 5’- TCCATCAACA-GCTTTAGCAGT -3’,[14]
BCL-XL forward, 5’- TGCATTGTTCCCATAGAGTTCCA -3’, and reverse, 5’- CCTGAATGACCACCTAGAGCCTT -3’,[20] and
GAPDH forward, 5’- GTCAACGGATTTGGTCGTATTG -3’, and reverse, 5’- CATGGGTGGAATCATATTGGAA -3’.[21]
qPCR was performed in StepOnePlus™ real time-PCR System (Applied Biosystems, USA). Triplicate PCRs (20 µl) were performed using SYBR™ Select Master Mix (Applied Biosystems, USA). The reaction mixture was initially set at 50°C for 2 min for UDG activation and then at 95°C for 2 min for AmpliTaq® DNA Polymerase, UP Activation and then subjected to 40 PCR cycles of 95°C for 3 s and 60°C for 30 s. A reference sample (K562 treated with Negative control [water]) was used to normalize data across experiments and mRNA levels were normalized to GAPDH levels.
Cell Proliferation Assay
Cell proliferation was determined using Cell Proliferation Kit I (3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide [MTT]) by Roche, Basel, Switzerland. The K562 was seeded in 24-well plates at a density of 2 × 105 cells/well. Each well was transfected with a required amount of siRNAs. Cultures were incubated at 37°C in a fully humidified atmosphere with 5% CO2. After 48 h of incubation, 60 µl of MTT (0.5 mg/ml) was added in each well and incubated for another 4 h. To dissolve the precipitate, 600 µl of dimethyl sulfoxide was then added to each well. The proliferation of K562 cells was quantified at an optical density of 490 nM. Cell viability was calculated as the percentage of amount of treated cells on control cells (cells treated with NT siRNA).
Apoptosis Detection Assay
The cell apoptotic rate was evaluated using a Fluorescein Isothiocyanate (FITC) Annexin V Apoptosis Detection Kit (FITC Annexin V Apoptosis Detection Kit I, BD Pharmingen, USA). The K562 cells in the logarithmic phase were collected, centrifuged at 1000 rpm for 5 min, and washed with precooled phosphate-buffered saline. ×1 binding buffer was added to the cell pellet to achieve a concentration of 1 × 106 cells/ml. 5 µl of FITC Annexin V and 5 µl propidium iodide (PI) were added to this cell suspension and incubated at RT for 15 min. 400 µl of ×1 binding buffer was added to each tube and analyzed by flow cytometry within 1 h.
Western Blot Analysis
Total proteins were extracted from the cells using NucleoSpin® RNA/Protein Kit by Macherey-Nagel GmbH & Co. (Germany) following the manufacturer’s protocol. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis was performed by loading 15 µg of protein in each lane. Proteins were then transferred to nitrocellulose membranes and then incubated with specific antibodies. The membranes were washed with Tris-buffered saline and Tween-20 and incubated with horseradish peroxidase (HRP)-conjugated second antibody for 1 h at room temperature (25°C). The blot was developed with SuperSignal™ West Pico PLUS Chemiluminescent Substrate, Thermo Scientific, USA, and the signal was exposed with X-ray film. All antibodies were from Thermo Scientific, USA and they were used in the following dilutions - GAPDH (1:5000), BCR-ABL (1:500), STAT5A (1:1000), STAT5B (1:1000), BCL-XL (1:250), and Goat anti-mouse immunoglobulin G (H+L) secondary antibody HRP (1:50,000).
Statistical Analysis
Results were expressed as mean ± standard deviation (SD) of at least three independent experiments performed in triplicate. GraphPad Prism 7 (GraphPad Software, Inc. CA, USA) was used for statistical analysis. The results were compared through one-way analysis of variance and differences between values were considered to be statistically significant at P < 0.05.
RESULTS
Confirmation of siRNA Transfection
siGLO Red siRNA was used to confirm the successful transfection into K562 cells. K562 cell was transfected with 50 nmol/l (nM) siGLO Red siRNA for 24 h. Further analysis was performed using fluorescent microscopy. From Figure 1, it can be confirmed that the siRNA has entered the cells and got localized near the nuclear region. The transfection efficiency was determined to be 69 ± 6.5% when 50 nM siRNA was used for 24 h by counting positively transfected cells per 100 cells in the field of view. HiPerFect transfection technique showed 9 ± 3.5% of cytotoxicity when compared with non-transfected cells.
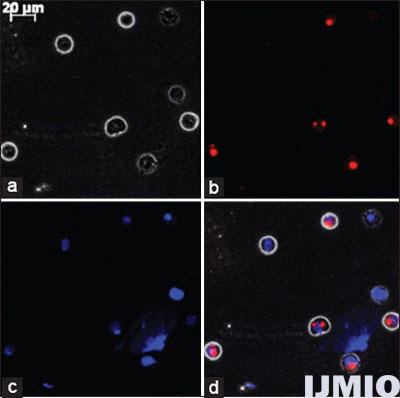
- Fluorescent microscopy image of K562 cells: K562 cell was transfected with 50 nM small interference (si) GLO Red siRNA for 24 h and observed under fluorescent microscopy at ×400 magnification. (a) Phase contrast image of K562. (b) siGLO siRNA seen as red dots. (c) Nuclear region stained in blue by DAPI. (d) Merge of all the three images
To determine whether 50 nM of each siRNA significantly reduced the gene expression in K562 cells, we transfected the cells with 50 nM of BCR-ABL, STAT5A, STAT5B, and BCL-XL siRNA individually for 48 h. After 48 h, the gene expression was detected through qPCR analysis and western blot analysis. The qPCR results showed that the mRNA level of each gene was significantly lowered in the transfected K562 cells than control cells (NT siRNA transfected K562 cells). The western blot results correlated with the qPCR results where the protein band intensity reduction was seen in siRNA-treated cells [Figure 2].
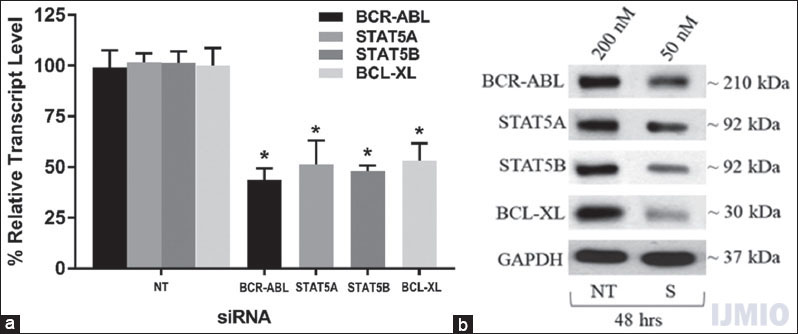
- Confirmation of small interference (si) RNA transfection: K562 was transfected with 50 nM each of BCR-ABL, STAT5A, STAT5B, and BCL-XL siRNA individually for 48 h. (a) Gene expression: The relative transcript level of each gene is expressed in percentage as compared to gene expression in control. GAPDH gene expression was used to normalize the expression of other target genes. Values are expressed in % ± SD; n = 3; *P < 0.05 when each value compared with their respective control. (b) Western Blot: With each sample, western blotting was carried out for the specific protein which was silenced with the respective siRNA. NT - K562 treated with 200 nM non-targeting siRNA, S - K562 cells treated with respective siRNAs at mentioned concentration and time point. The blot shown is a representative figure of three individual experiments
Effect of Silencing Single Gene
First, we wanted to study the effect of each siRNA on the gene expressions, cell proliferation, and apoptosis of K562. For this purpose, 50 nM of each siRNA was transfected into K562 individually, and gene expression of all four genes was studied after 48 h. When BCR-ABL and STAT5A genes were silenced individually, the BCL-XL expression was lowered than the control sample by around 10%. Similarly, when STAT5A and BCL-XL gene was suppressed, the expression of BCR-ABL was lowered as compared to the control by approximately 10%. None of the above-mentioned difference was statistically significant, except when the target gene suppression was determined for each of the siRNA used [Figure 3a]. MTT test results revealed that the cell proliferation rate of the transfected cells was lower than that of the control cells. The lowest proliferation rate was observed when BCL-XL siRNA was used and it was found to be 66.34 ± 9.7% [Figure 3b].
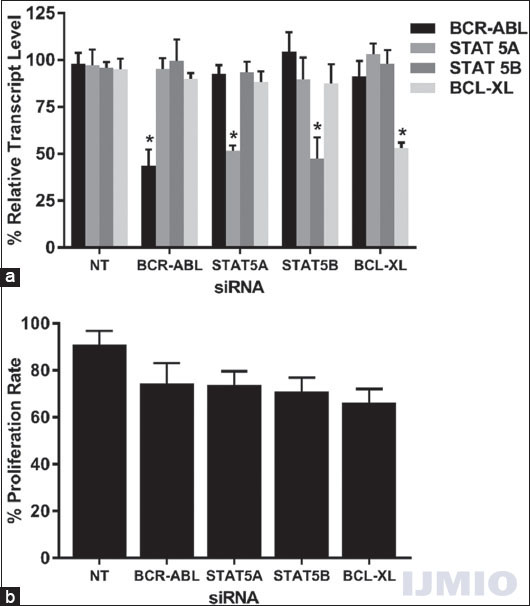
- Effect of silencing single gene: K562 was transfected with 50 nM each of BCR-ABL, STAT5A, STAT5B, and BCL-XL siRNA individually for 48 h. (a) Gene expression: The relative transcript level of each gene is expressed in % as compared to gene expression in control. GAPDH gene expression was used to normalize the expression of other target genes. Values are expressed in % ± SD; n = 3; *P < 0.05 when each value compared with their respective control. (b) Cell proliferation: K562 proliferation rate is expressed in % as compared to the proliferation rate in control. Values are expressed in % ± SD; n = 3; *P < 0.05 when compared with control
In the previous experiments, all the four siRNAs could inhibit K562 cells proliferation to some extent. To investigate whether these silencing of genes accelerates K562 cells apoptosis, Annexin V/PI staining was carried out followed by flow cytometry analysis. Compared to the control cells, the apoptotic cells increased highest by 24 ± 13.7% in BCL-XL silenced cell population and lowest by 20 ± 9.1% in BCR-ABL silenced cell population. None of the results were statistically significant when compared with control cells. However, these results indicated that silencing BCR-ABL, STAT5A, STAT5B, and BCL-XL individually induced K562 cells to undergo apoptosis [Figure 4].
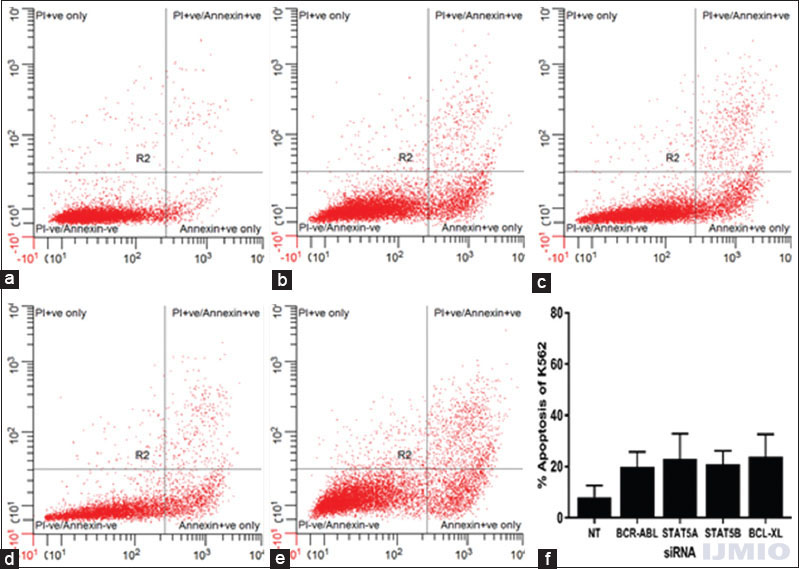
- Apoptotic status after individual gene silencing: K562 cells were assayed through flow cytometry using Annexin V/PI staining. Data are representative of three separate independent experiments. (a) - With 200 nM of NT siRNA. (b) With 50 nM BCR-ABL small interference (si) RNA. (c) With 50 nM STAT5A siRNA. (d) With 50 nM STAT5B siRNA. (e) With 50 nM BCL-XL siRNA. (f) Apoptotic rate is expressed in % of Annexin V-positive events in a total number of events. Values are expressed in % ± SD; n = 3; *P < 0.05 when compared with control
Effect of Silencing Two Genes
Second, we intended to study the effect of silencing two genes at a time in STAT pathway in K562. Since we had four target genes, a total number of combinations comprising two different siRNAs were six. Each siRNA was used at 50 nM concentration and the cells were incubated for 48 h. Every siRNA significantly reduced its target gene’s expression by more than 50% when compared with the NT siRNA-treated control. It was observed that, in some instances, simultaneous treatment with two siRNAs showed improved effects in silencing of its target genes. For example, the gene silencing improved when BCR-ABL and STAT5A were silenced together (40.95 ± 14.5% and 45.58 ± 11.01%) as compared to when they were silenced individually [43.63 ± 13.7% and 51.54 ± 5.4%, Figure 3a]. Furthermore, BCL-XL showed improved silencing in all combination of siRNA [Figure 5a]. The cell proliferation rate was seen to be unambiguously reduced in all double siRNA combinations as compared to single siRNA application. Statistically significant reduction in cell proliferation was seen in two occasions: one when BCR-ABL and STAT5A siRNA were used together (57.8 ± 10.8%) and another when BCR-ABL and BCL-XL siRNA were used together (59.5 ± 8.2%) [Figure 5b]. More cells underwent apoptosis when two siRNAs were used together, and the highest apoptosis rate (39.74 ± 9.4%) was seen when BCR-ABL and BCL-XL siRNA were used together [Figure 5c].
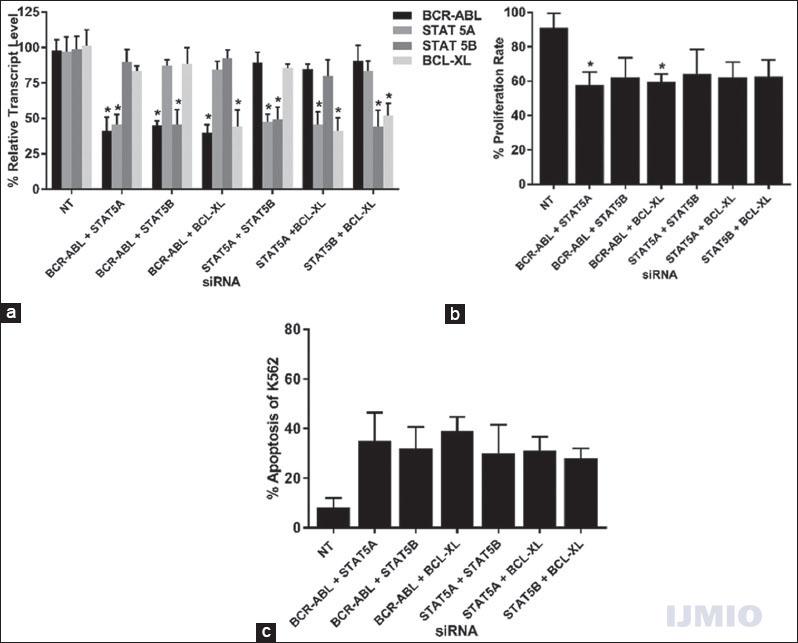
- Effect of silencing two genes: K562 cells were transfected with different combinations of BCR-ABL, STAT5A, STAT5B, or BCL-XL siRNAs at 50 nM each for 48 h. (a) Gene expression: The relative transcript level of each gene is expressed in % as compared to gene expression in control. GAPDH gene expression was used to normalize the expression of other target genes. Values are expressed in % ± SD; n = 3; *P < 0.05 when each value compared with their respective control. (b) Cell proliferation: K562 proliferation rate is expressed in % as compared to the proliferation rate in control. Values are expressed in % ± SD; n = 3; *P < 0.05 when compared with control. (c) Apoptosis: K562 apoptotic rate is expressed in % of Annexin V-positive events in a total number of events. Values are expressed in % ± SD; n = 3; *P < 0.05 when compared with control
Effect of Silencing Three Genes
The penultimate silencing strategy was to silence any three genes simultaneously and study its effect of gene silencing, cell proliferation, and apoptosis. In this category, four combinations of siRNA were possible with a total concentration of 150 nM for every combination. Every siRNA significantly reduced its target gene in all combination used. The non-targeted gene in every combination also showed reduced gene expression. The highest non-targeted reduction was seen in BCL-XL gene expression with 74.64 ± 13.6% when the other three genes were silenced [Figure 6a]. The cell proliferation in each combination was significantly reduced when compared with the NT siRNA-transfected control [Figure 6b]. The apoptotic rate of K562 increased significantly when three siRNAs were used together. In both the instances, the highest reduction in cell proliferation and an increase in apoptotic rate were seen when BCR-ABL siRNA was used along with STAT5A and BCL-XL siRNA [Figure 6c].
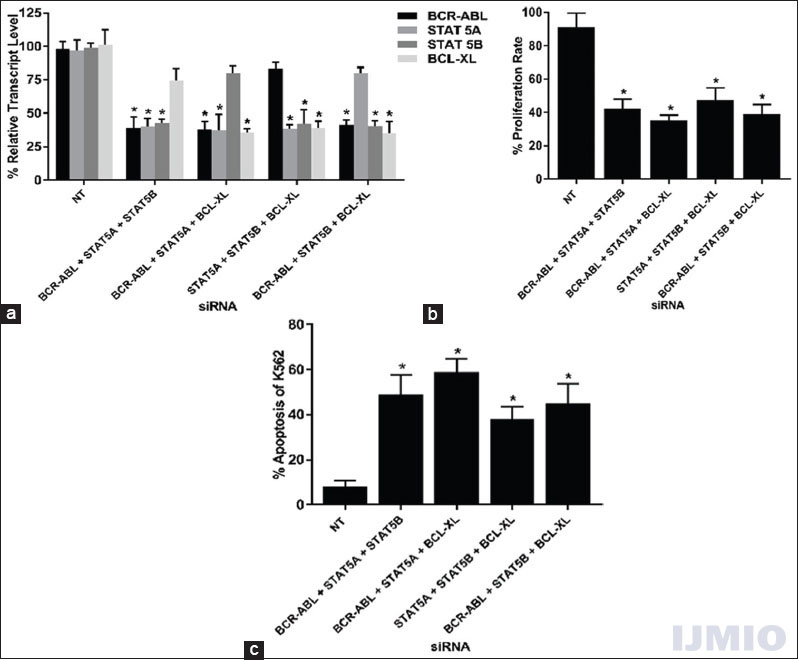
- Effect of silencing three genes: K562 cells were transfected with different combinations of BCR-ABL, STAT5A, STAT5B, or BCL-XL siRNAs at 50 nM each for 48 h. (a) Gene expression: The relative transcript level of each gene is expressed in % as compared to gene expression in control. GAPDH gene expression was used to normalize the expression of other target genes. Values are expressed in % ± SD; n = 3; *P < 0.05 when each value compared with their respective control. (b) Cell proliferation: K562 proliferation rate is expressed in % as compared to the proliferation rate in control. Values are expressed in % ± SD; n = 3; *P < 0.05 when compared with control. (c) Apoptosis: K562 apoptotic rate is expressed in % of Annexin V-positive events in a total number of events. Values are expressed in % ± SD; n = 3; *P < 0.05 when compared with control
Effect of Silencing Four Genes
Finally, as a therapeutic approach, all four genes were silenced simultaneously with 50 nM concentration of each siRNA. The gene expression of all target genes was significantly reduced when compared with the control. When all four genes were silenced simultaneously, the gene expression of BCL-XL was reduced the most and STAT5B was reduced the least. The cell proliferation rate of K562 was reduced by around 75% and the apoptosis was induced in more than 60% of K562 cells when all four siRNAs were used together [Table 1].
| siRNA used | A. % mRNA remaining | B. Cell proliferation rate (%) | C. apoptosis rate (%) | |||
|---|---|---|---|---|---|---|
| BCRABL | STAT5A | STAT5B | BCLXL | |||
| NT | 98.32±9.64 | 97.85±15.67 | 99.21±14.92 | 101.78±9.87 | 91.05±13.34 | 8.36±3.47 |
| BCRABL+STAT5A+STAT5B+BCLXL | 33.01±10.2 | 36.94±9.81 | 40.77±12.93 | 31.17±11.63 | 25.21±9.25 | 64.15±10.47 |
K562 cells were transfected with BCRABL, STAT5A, STAT5B, and BCLXL siRNAs together at 50 nM each for 48 h. A: Gene expression: The relative transcript level of each gene is expressed in %as compared to gene expression in control. GAPDH gene expression was used to normalize the expression of other target genes. Values are expressed in %±SD; n=3; Bold values P<0.05 when each value compared with their respective control. B: Cell proliferation: Proliferation rate are expressed in % as compared to the proliferation rate in K562 cells treated with NT siRNA (control). Values are expressed in %±SD; n=3; Bold values P<0.05 when compared with Control. C: Apoptotic status: Apoptotic rate are expressed in % of Annexin Vpositive events in a total number of events. Values are expressed in %±SD; n=3; Bold values P<0.05 when compared with control
Multiple Gene Silencing is Better to Individual Gene Silencing
It was evident from all the previous experiments that silencing BCR-ABL, STAT5A, STAT5B, and BCL-XL simultaneously was the most efficient way to kill K562 cells in this setup. We wanted to confirm that this was an additive effect of siRNAs and not merely a cause of higher concentration of siRNAs. To prove this, we individually transfected high concentration (200 nM) of each siRNA in K562 cells and compared the results with all 4 siRNA-transfected cells. Each siRNA at 200 nM concentration reduced its target gene to a range of 9–14%, while in all 4 siRNAs, silencing the genes was reduced to a range of 31–41% [Figure 7a]. High concentration of individual siRNA was not able to reduce cell proliferation to >41.5%, but all four siRNAs were able to reduce the cell proliferation rate to 25.21 ± 9.2% [Figure 7b]. Similarly, the highest rate of apoptosis (51.25 ± 14.97%) was induced when BCL-XL siRNA was used at 200 nM, but all four siRNAs were able to induce 64.15 ± 10.4% apoptosis in K562 cells [Figure 7c].

- Comparison of individual silencing with high concentration small interference (siRNA) and simultaneous gene silencing. K562 cells were transfected individually with 200 nM of BCR-ABL, STAT5A, STAT5B, and BCL-XL siRNA and all 4 siRNAs with 50 nM of each siRNA together for 48 h. (a) Gene expression: The relative transcript level of each gene is expressed in % as compared to gene expression in Control. GAPDH gene expression was used to normalize the expression of other target genes. Values are expressed in % ± SD; n = 3; *P < 0.05 when each value compared with their respective control. (b) Cell proliferation: K562 proliferation rate is expressed in % as compared to the proliferation rate in control. Values are expressed in % ± SD; n = 3; *P < 0.05 when compared with control. (c) Apoptosis: K562 apoptotic rate is expressed in % of Annexin V-positive events in a total number of events. Values are expressed in % ± SD; n = 3; *P < 0.05 when compared with control
DISCUSSION
In CML, BCR-ABL displays constitutive phosphorylation activity of tyrosine residues in various cellular proteins.[22] Although STAT5A and STAT5B share a very similar structure, they have different functions in CML. STAT5 and AKT together drive the oncogenesis in CML.[23] BCL-XL plays a very important role in cell survival and inhibits apoptosis in K562 cells.[15] This study investigated the effect of silencing BCR-ABL, STAT5A, STAT5B, and BCL-XL individually and simultaneously in different combinations. Gene expression profile, cell proliferation rate, and apoptosis induction rate were studied after transfection with siRNA/s. We could demonstrate that multiple siRNA treatments for K562 cells were highly effective as compared to single siRNA treatment for 48 h with 50 nM concentration of each siRNA.
Individually, 50 nM concentration of siRNA decreased mRNA, reduced cell proliferation, and increased apoptosis in K562 cells [Figures 3 and 4]. While single target inhibition showed only slight effects, the combined silencing caused a statistically significant decrease in cell proliferation and significantly increased the apoptosis rate in K562 cells [Table 1]. Simultaneous gene silencing was also the most efficient combination to inhibit cell proliferation and induce apoptosis compared to all other combinations of siRNAs [Figures 5 and 6]. We were also able to demonstrate the non-target silencing effect where the BCL-XL expression was reduced when STAT5A and STAT5B were silenced together [Figure 5] similar to a work by Yanli et al.[24]
Targeting four genes in the STAT pathway was more effective in triggering apoptosis and inhibition of cell proliferation when compared to the silencing of three or less number of genes. This could be because, when a single gene is blocked, the cell switches to alternate pathways for survival. However, when a more number of pathway-related genes are silenced simultaneously, it becomes difficult for the cells to substitute for each blocked protein. Hence, simultaneous gene silencing blocks signaling pathways with or without additional targeting of the key regulatory gene/s. The basic toxicity of siRNAs is well studied and documented. Lowering the concentration of each siRNA would reduce the off-target effects and also avoids the RNAi endogenous pathway to get saturated with a single type of siRNA in a cell.[25] We were able to show that using all 4 siRNAs at 50 nM concentration each was much more effective than using any single siRNA at 200 nM concentration [Figure 7].
Certain shortcomings must be highlighted in the reported experiments. K562 cell line was the only cell model used in this study; we restricted the studies to this model due to its close comparison with CML disorder. Another limitation might be that the present study focused on only one of the many pathways in CML. Simultaneous gene silencing targeting key players in other pathways of CML would be an interesting approach in this kind of the study. Finally, in vivo experiment would have been helpful in ascertaining our findings in K562.
CONCLUSIONS
We were able to show that, in K562 cells, simultaneous silencing of BCR-ABL, STAT5A, STAT5B, and BCL-XL was more efficient in inhibiting cell proliferation and inducing apoptosis than silencing these genes individually or in any other combinations.
REFERENCES
- Tyrosine kinase inhibitors in acute and chronic leukemias. Expert Opin Pharmacother. 2012;13:927-38.
- [Google Scholar]
- Clinical and prognostic significance of e1a2 BCR-ABL1 transcript subtype in chronic myeloid leukemia. Blood Cancer J. 2017;7:e583.
- [Google Scholar]
- Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of BCR-ABL positive cells. Nat Med. 1996;2:561-6.
- [Google Scholar]
- T315I, more or less, predicts for major molecular response: The devil is in the details! Haematologica. 2013;98:665-6.
- [Google Scholar]
- Current approach to the treatment of chronic myeloid leukaemia. Leuk Res. 2017;55:65-78.
- [Google Scholar]
- Past, present, and future of BCR-ABL inhibitors: From chemical development to clinical efficacy. J Hematol Oncol. 2018;11:84.
- [Google Scholar]
- Hallmarks of cancer: The next generation. Cell. 2011;144:646-74.
- [CrossRef] [PubMed] [Google Scholar]
- Systemic review on chronic myeloid leukemia: Therapeutic targets, pathways and inhibitors. J Nucl Med Radiat Ther. 2015;6:257.
- [Google Scholar]
- BCR-ABL uncouples canonical JAK2-STAT5 signaling in chronic myeloid leukemia. Nat Chem Biol. 2012;8:285-93.
- [Google Scholar]
- Blockade of the BCR-ABL kinase activity induces apoptosis of chronic myelogenous leukemia cells by suppressing signal transducer and activator of transcription 5-dependent expression of bcl-xL. J Exp Med. 2000;191:977-84.
- [Google Scholar]
- Therapeutic miRNA and siRNA: Moving from bench to clinic as next generation medicine. Mol Ther Nucleic Acids. 2017;8:132-43.
- [Google Scholar]
- Specific inhibition of BCR-ABL gene expression by small interfering RNA. Blood. 2003;101:1566-9.
- [Google Scholar]
- Suppression of STAT5A and STAT5B chronic myeloid leukemia cells via siRNA and antisense-oligonucleotide applications with the induction of apoptosis. Am J Blood Res. 2013;3:58-70.
- [Google Scholar]
- Bcl-xL is a dominant antiapoptotic protein that inhibits homoharringtonine-induced apoptosis in leukemia cells. Mol Pharmacol. 2011;79:1072-83.
- [Google Scholar]
- Progress in RNAi-mediated molecular therapy of acute and chronic myeloid leukemia. Mol Ther Nucleic Acids. 2015;4:e240.
- [Google Scholar]
- Additive antileukemia effects by GFI1B- and BCR-ABL-specific siRNA in advanced phase chronic myeloid leukemic cells. Cancer Gene Ther. 2013;20:421-7.
- [Google Scholar]
- Single and combinational siRNA therapy of cancer cells: Probing changes in targeted and non-targeted mediators after siRNA treatment. Mol Pharm. 2016;13:4116-28.
- [Google Scholar]
- BCR-ABL/GATA1/miR-138 mini circuitry contributes to the leukemogenesis of chronic myeloid leukemia. Oncogene. 2014;33:44-54.
- [Google Scholar]
- Quinacrine induces apoptosis in human leukemia K562 cells via p38 MAPK-elicited BCL2 down-regulation and suppression of ERK/c-jun-mediated BCL2L1 expression. Toxicol Appl Pharmacol. 2015;284:33-41.
- [Google Scholar]
- Dual small-molecule targeting of SMAD signaling stimulates human induced pluripotent stem cells toward neural lineages. PLoS One. 2014;9:e106952.
- [Google Scholar]
- Structure, regulation, signaling, and targeting of Abl kinases in cancer. Genes Cancer. 2012;3:436-46.
- [Google Scholar]
- Co-operating STAT5 and AKT signaling pathways in chronic myeloid leukemia and mastocytosis: Possible new targets of therapy. Haematologica. 2014;99:417-29.
- [Google Scholar]
- Targeting methyltransferase PRMT5 eliminates leukemia stem cells in chronic myelogenous leukemia. J Clin Invest. 2016;126:3961-80.
- [Google Scholar]
- Oncogene dependency and the potential of targeted RNAi-based anti-cancer therapy. Biochem J. 2014;461:1-3.
- [Google Scholar]






